Installing Packages and Bioconductor Overview
Overview
Teaching: 20 min
Exercises: 5 minQuestions
What is CRAN?
How do I install packages?
What is Bioconductor?
How do I install Bioconductor packages?
How do I find help in Bioconductor?
Objectives
Manage packages
Manage Bioconductor Packages
Navigate Bioconductor
Gain access to Bioconductor
Learn about packages to access data
Other output formats
R Packages and CRAN
It is possible to add functions to R by writing a package, or by obtaining a package written by someone else. One of the primary ways in which packages are distributed is through centralized repositories. The first R repository a user typically runs into is the Comprehensive R Archive Network (CRAN), As of this writing, there are over 17,000 packages available on CRAN, the home of many of the most popular R packages. R and RStudio provide functionality for managing packages:
From the console:
- You can see what packages are installed by typing
installed.packages() - You can install packages by typing
install.packages("packagename"), wherepackagenameis the package name, in quotes. - You can update installed packages by typing
update.packages() - You can remove a package with
remove.packages("packagename") - You can make a package available for use with
library(packagename)
Using the RStudio interface
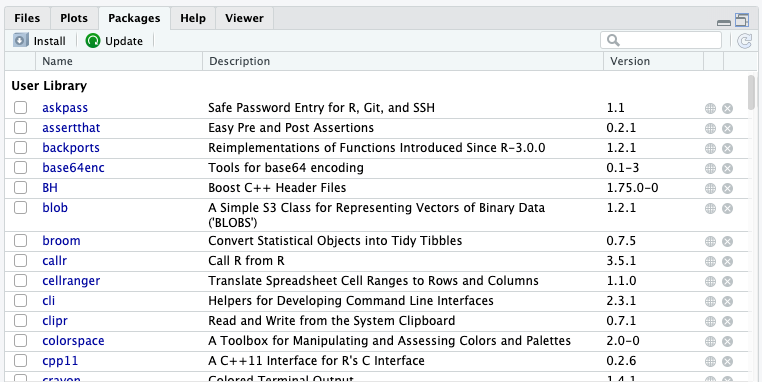
You can also use the RStudio interface to view and install packages. Pane 4 (which might be different for you if you’ve personalized RStudio’s interface) provides a Packages tab that allows you to see the packages you have installed and loaded at a given point in time. The Packages tab is broken down into your User Library, these are the packages you have installed throughout your R use experience, and also the System Library the packages that are part of the R kernel which is updated when you update your version of R.
The default packages interface for RStudio:
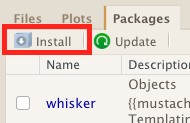
To install a package click Install
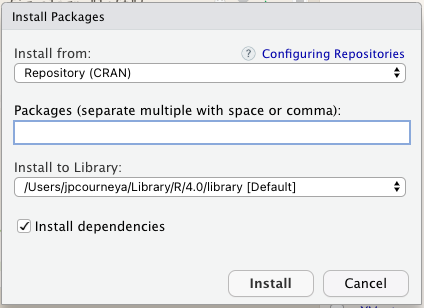
In the pop-up window type the package name of interest.
Loading Packages
When it comes time to load a package you have installed it can also be done a number of ways most commonly it will be done using the console or writing it at the beginning of a script since the first thing you should be doing in your script is loading libraries.
From the console:
library(<package>)
Using the RStudio interface:
 Click the white box next to a package in your library and that will load the library in to your session.
Click the white box next to a package in your library and that will load the library in to your session.
Tip: Beware of loading conflicts
Pay attention to the messages that are being printed to the console when you load a package. Sometimes you will see
The following objects are masked from.... This is telling you that when the package is loaded the function that typically is related to another package is now being referenced by the package you loaded. Sometimes this can cause a conflict between other packages which depend on the native function being masked and cause your code to break. Beware!
Exercise: Install and Load a package from CRAN
Using both the console and the RStudio interface install and load the
ggplot2package with the console anddplyrpackage with the RStudio interface.Solution
From the console:
install.packages("ggplot2") #installs ggplot2 library(ggplot2) #loads the packageUsing the RStudio interface:
Using the
Packagestab inPane 4click on Install button and type indplyr. Next proceed to click the Check Box next todplyrto load it.
About Biocondutor
Similar to CRAN, Bioconductor is a repository of R packages as well. Bioconductor provides tools for the analysis and comprehension of high-throughput genomic data. Bioconductor uses the R statistical programming language, and is open source and open development. It has two releases each year, and an active user community.
Installing Bioconductor
In order to install Bioconductor packages you first need the BiocManager package which is hosted on CRAN. To install it you will need to run:
install.packages("BiocManager")
Bioconductor releases and current version
The Bioconductor project produces two releases each year, one around April and another one around October.
The April release of Bioconductor coincides with the annual release of R. Packages in that Bioconductor release are tested for the upcoming version of R. Users must install the new version of R to access the new version of those packages.
The October release of Bioconductor continue to work with the same version of R for that annual cycle.
Each time a new release is made, the minor version of all the packages in the Bioconductor repository is incremented by one.
Once the BiocManager package is installed, the BiocManager::version() function displays the version (i.e., release) of the Bioconductor project that is currently active in the R session.
BiocManager::version()
[1] '3.12'
Installing Bioconductor packages
The BiocManager::install() function is used to install packages.
The function first searches for the requested package(s) on the Bioconductor repository, but falls back on the CRAN repository and also supports installation from GitHub repositories. There is a lengthy explanation by Bioconductor maintainers as to why using this is prefered over install.packages(). Find that here
We can install the BiocPkgTools package which provides a collection of simple tools for learning about Bioc Packages we’d install it as so:
BiocManager::install("BiocPkgTools")
Explore the package universe
library(BiocPkgTools) #loads BiocPkgTools
biocExplore() #interactive visualization of package info.
Check for updates
The BiocManager::valid() function checks the version of currently installed packages, and checks whether a new version is available for any of them on the Bioconductor repository.
Conveniently, if any package can be updated, the function generates and displays the command needed to update those packages. Users simply need to copy-paste and run that command in their R console.
If everything is up-to-date, the function will simply print TRUE.
BiocManager::valid()
Bioconductor Help
Bioconductor stands apart from CRAN in that it requires packages to have documentation available and workflows. Bioconductor also provides a Bioconductor specific support site that is a Stack Overflow type experience. The site is possible due to the contribution of developers in the Bioconductor community as well as countless dedicated volunteers answering questions.
Bioconductor Packages
Each package hosted on Bioconductor has a dedicated page with various resources. For an example, looking at the scater package page on Bioconductor, we see that it contains:
-
a brief description of the package at the top, in addition to the authors, maintainer, and an associated citation
-
installation instructions that can be cut and paste into your R console
-
documentation - vignettes, reference manual, news
You can gain the most traction using a package by looking at its documentation section.
-
Each Bioconductor package contains at least one vignette, a document that provides a task-oriented description of package functionality. Vignettes contain executable examples and are intended to be used interactively.
-
The reference manual contains a comprehensive listing of all the functions available in the package. This is a compilation of each function’s manual, aka help pages, which can be accessed programmatically in the R console via ?
. -
Finally, the NEWS file contains notes from the authors which highlight changes across different versions of the package. This is a great way of tracking changes, especially functions that are added, removed, or deprecated, in order to keep your scripts current with new versions of dependent packages.
Below this, the Details section covers finer nuances of the package, mostly relating to its relationship to other packages:
-
upstream dependencies (Depends, Imports, Suggests fields): packages that are imported upon loading the given package
-
downstream dependencies (Depends On Me, Imports Me, Suggests Me): packages that import the given package when loaded
For example, we can see that an entry called simpleSingle in the Suggests Me field on the scater page takes us to a step-by-step workflow for low-level analysis of single-cell RNA-seq data.
BiocViews
One additional Details entry, the biocViews, is helpful for looking at how the authors annotate their package. For example, for the scater package, we see that it is associated with DataImport, DimensionReduction, GeneExpression, RNASeq, and SingleCell, to name but some of its many annotations.
The BiocViews page provides a hierarchically organized view of annotations associated with Bioconductor packages. Under the “Software” label for example (which is comprised of most of the Bioconductor packages), there exist many different views to explore packages. For example, we can inspect based on the associated “Technology”, and explore “Sequencing” associated packages, and furthermore subset based on “RNASeq”.
Another area of particular interest is the “Workflow” view, which provides Bioconductor packages that illustrate an analytical workflow. For example, the “SingleCellWorkflow” contains the aforementioned tutorial, encapsulated in the simpleSingleCell package.
Accessing Publicly Available Data
The NCBI Gene Expression Omnibus (GEO) is a public repository of microarray data. Given the rich and varied nature of this resource, it is only natural to want to apply BioConductor tools to these data. GEOquery is the bridge between GEO and BioConductor.
Getting data from GEO is really quite easy. There is only one command that is needed, getGEO. This one function interprets its input to determine how to get the data from GEO and then parse the data into useful R data structures. Usage is quite simple. After installing GEOquery BiocManager::install("GEOquery"), loads the GEOquery library:
library(GEOquery)
Now, you are free to access any GEO accession. In general, you will use only the GEO accession.
gds <- getGEO("GDS507") #provide a GEO Accession ID and assign it to an object.
To learn more take advantage of the GEOquery vignette:
browseVignettes(package = "GEOquery")
Key Points
Both CRAN and Bioconductor provide a pantheon of packages to extend R.
Use
install.packages()to install packages (libraries).
BiocManager::install()is the recommended way to install Bioconductor packages.